I have developed a software in Windows 10. It simulates molecular markers and genes. I am generating a data set of 4900 rows x 60000 columns in two equivalent Dell Optiplex. My Optiplex 7070 has Windows 10, intel i7-9th generation, 32 gb of RAM. My Optiplex 5090 has Ubuntu 20, intel i7-10th generation, 32 gb of RAM. Why do my software is much, much lower in Ubuntu than in Windows? Both version of my software are 64 bits, compiled using Xojo 2022r1. Any information on how to make my software faster in Ubuntu?
Is the speed difference in the manipulation of the data, or in the visual representation of it afterwards?
Only Xojo can tell you if the compiled code differs in any great extent.
If the data is made up from 64bit integers, this array will occupy over 1 Gb or memory.
If it is bigger, then you could be into virtual memory territory
What is the data?
Does it all need to be in memory at once?
What does ‘simulation’ entail?
It’s been a while since I did Linux code in Xojo, but I remember there being some differences in threaded performance.
Are you using #pragma BackgroundTasks false consistently in tight loops? It’s very important. (Some of the other #pragma directives such as BoundsChecking, NilObjectChecking, and StackOverflowChecking can also have a big impact)
Dear Jeff,
It is not simply to explain but there are lots of processing before generating a file of, for example, 4900 individuals x 60000 SNPs (a DNA marker, coded 11, 12, and 22). Because I code only in the Windows environment, the code executed by Linux is exactly the same. I used regularly threads, to give the user the opportunity to work in other programs. To finished the process, including saving the files, windows expended 24 hours while Linux expended almost 10 days. It is unbelievable! I really keeps most information in RAM but I noticed the Linux used less than 2% of the free memory. I will check this in Windows. But I never had problems of memory in my Windows version. I really need help because I cannot offer my software to Linux users (scientists, professors, and graduate students in the areas of human genetics and animal and plant genetics and breeding).
Marcelo
Dear Mike,
Please, see my reply to Jeff. Should I use specific commands to Linux? Where I can find such information for coding for Linux environment? I am a costumer of Xojo Developer but I did not had time to read the magazine over the last years.
Marcelo
That is unbelieveable.
Have you looked at where the bulk of the time is spent?
You mention ‘including saving the file’ … thats an alarm bell.
Does that take a long time?
Maybe the hard drive is damaged or very fragmented.
Or is it mostly the calculations?
It’s almost impossible to say how this can be modified or optimised without seeing at least some of the code.
If you would like to privately send me a copy (I promise it is safe) , I might be able to suggest some tweaks.
Dear Jeff,
No problem to send the thread. Linux is spending days processing a thread. There are lots of loops in this thread. Saving in Linux is as fast as in Windows. However, only in the next Monday I can send the code. Now I am traveling and out of my university (Federal University of Vicosa).
Marcelo
No worries. When youre ready.
Dear Jeff,
I requested to one of my graduate students to send the thread code to me by email. When I get it I will share with you. I think that you will suggest using pragma directives as Mike did. Regards,
Marcelo
and other things
On macOS, it’s very easy to Sample a running process and to show the time spent in each thread with a stack trace. (You can do this in macOS using Activity Monitor.app, or for more control and detail use Instruments.app). There must be a similar feature available on Linux?
Dear Jeff,
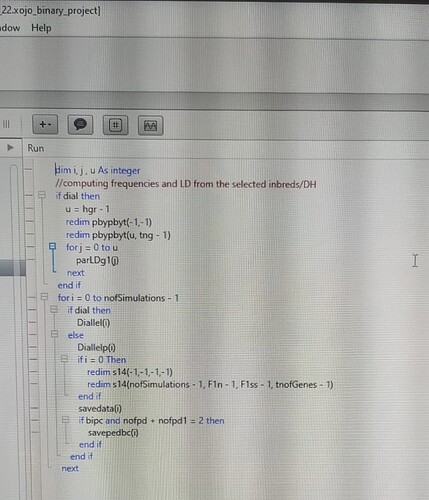
My software (REALbreeding) has several classes, modules, and windows. There are several threads too. After preparing a Linux version, from my Windows version, I choose a procedure called Diallel to compare both versions. See in the first picture how I call the thread:
The thread has only few lines but a method that demands high processing:
The method used for the test of relative processing time is Diallel. The code for this method is below.
dim u1, sl, u2, i, j, cc, k, idx1, idx2, sl1, il As integer, s As String, ihg1, ihg2, tGen(), tGen1() As int8, sel1, sel2 As Boolean
reds
if idxs = 0 then
f1Id = “”
end if
if dComplete then
if not csIl then
u1 = (tnofpar*(tnofpar - 1)/2) - 1
else
u1 = (tnofspar*(tnofspar - 1)/2) - 1
end if
else
i = 0
nf1pd = 0
for j = 1 to hgr - 1
if not csIl then
nf1pd = nf1pd + nilhg(i)*nilhg(j)
else
nf1pd = nf1pd + nsilhg(i)*nsilhg(j)
end if
next
u1 = nf1pd - 1
end if
if dComplete then
sl = tnofpar - 2
sl1 = sl + 1
else
sl = nilhg(0) - 1
sl1 = nilhg(1) - 1
end if
//Chr 1
u2 = rnmbyChr(0) - 1
redim indgenG12(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG13(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g1positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG13(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g1positions)
for k = 0 to u2
indgenG12(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
if idxs = 0 then
if cc - 1 < u1 then
s = “;”
else
s = “”
end if
f1Id = f1Id + cstr(ihg1 + 1) + " " + cstr(ihg2 + 1) + " " + cstr(idx1 + 1) + " " + cstr(idx2 + 1) + s
end if
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
if chrnumber > 1 then
//Chr 2
u2 = rnmbyChr(1) - 1
redim indgenG22(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG23(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g2positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG23(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g2positions)
for k = 0 to u2
indgenG22(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
end if
if chrnumber > 2 then
//Chr 3
u2 = rnmbyChr(2) - 1
redim indgenG32(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG33(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g3positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG33(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g3positions)
for k = 0 to u2
indgenG32(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
end if
if chrnumber > 3 then
//Chr 4
u2 = rnmbyChr(3) - 1
redim indgenG42(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG43(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g4positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG43(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g4positions)
for k = 0 to u2
indgenG42(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
end if
if chrnumber > 4 then
//Chr 5
u2 = rnmbyChr(4) - 1
redim indgenG52(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG53(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g5positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG53(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g5positions)
for k = 0 to u2
indgenG52(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
end if
if chrnumber > 5 then
//Chr 6
u2 = rnmbyChr(5) - 1
redim indgenG62(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG63(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g6positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG63(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g6positions)
for k = 0 to u2
indgenG62(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
end if
if chrnumber > 6 then
//Chr 7
u2 = rnmbyChr(6) - 1
redim indgenG72(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG73(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g7positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG73(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g7positions)
for k = 0 to u2
indgenG72(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
end if
if chrnumber > 7 then
//Chr 8
u2 = rnmbyChr(7) - 1
redim indgenG82(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG83(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g8positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG83(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g8positions)
for k = 0 to u2
indgenG82(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
end if
if chrnumber > 8 then
//Chr 9
u2 = rnmbyChr(8) - 1
redim indgenG92(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG93(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g9positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG93(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g9positions)
for k = 0 to u2
indgenG92(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
end if
if chrnumber > 9 then
//Chr 10
u2 = rnmbyChr(9) - 1
redim indgenG102(0, idxs, u1, u2)
for i = 0 to sl
if dComplete then
ihg1 = hgId(i + 1)
else
ihg1 = 0
end if
if ihg1 = 0 then
idx1 = i
else
idx1 = parid(ihg1, i)
end if
redim tgam(-1)
redim tgam(u2)
redim tgen(-1)
redim tgen(u2)
//feminine gamete
if csIl then
if all(ihg1) then
sel1 = true
else
sel1 = checkil(idxs, ihg1, idx1)
end if
end if
if not csIl or (csIl and sel1) then
for k = 0 to u2
tgen(k) = indgenG103(ihg1, idxs, idx1, k)
next
gamfromGen(tgam, tgen, g10positions)
if dComplete then
il = i + 1
else
il = 0
end if
for j = il to sl1
if dComplete then
ihg2 = hgId(j + 1)
else
ihg2 = 1
end if
if dComplete or ihg1 <> ihg2 then
if ihg2 = 0 or not dComplete then
idx2 = j
else
idx2 = parid(ihg2, j)
end if
redim tgam1(-1)
redim tgam1(u2)
redim tgen1(-1)
redim tgen1(u2)
//masculine gamete
if csIl then
if all(ihg2) then
sel2 = true
else
sel2 = checkil(idxs, ihg2, idx2)
end if
end if
if not csIl or (csIl and sel2) then
cc = cc + 1
for k = 0 to u2
tgen1(k) = indgenG103(ihg2, idxs, idx2, k)
next
gamfromGen(tgam1, tgen1, g10positions)
for k = 0 to u2
indgenG102(0, idxs, cc - 1, k) = cdbl(tgam(k) + tgam1(k))
next
end if
end if
if csIl and cc = u1 + 1 then
exit
end if
next
end if
if csIl and cc = u1 + 1 then
exit
end if
next
cc = 0
end if
if idxs = 0 then
redim s11(nofSimulations - 1, u1, tnofGenes - 1)
end if
savedata(idxs)
Please, remember that my Windows version is very fast. But I really need to develop a version to Linux and for iOS (my next problem). I really need some help not only to speed up this method, but all methods in my software. And there are many!
Thanks for your attention. Regards,
Marcelo
You show
da.run
while da..State = 0
// wait
wend
What are you doing to “wait”? If you’re not sleeping the main thread while you “wait”, you’re going to starve the da thread for time. That just makes everything slower. And that could be really bad on Linux, as @Mike_D suggested.
Is this one method? Holy guacamole!!!
- Give your variables better names.
- My rule of thumb is to have a method visible on the screen completely.
Shows all the signs of having been translated from a different language/ different Basic
To me, a lot of time is likely being wasted with the redims
It should be possible to keep a constant (large enough) array available, and just make a note about how much of it is used.
In simple terms, if you have a college with a potential 2000 stutends, your array could start at 2000.
You record number_of_students - sometimes it is 1000, sometimes it is 1700, but the array never changes.
That does mean there can be unused data ‘above the number’, but as long as you stop at the right spot, you will be fine.
Not redimming means no constant reallocation of memory.
Without comments, it is really hard to follow what this code does - it looks like it has grown organically, with IF statements added to cater for edge cases during testing.
The code shown is not optimised… adding the #pragma statements would help, but the truly slow part could be in any of the methods that this calls.
eg gamfromGen
Maybe the SaveData part can be optimised… each time that is called, you might open a file on disc and write to it.
Changing that to ‘add a line to an in-memory string’, then writing the whole lot at the end might improve things, depending upon the size of the final data file.
my Windows version is very fast.
It’s not. It’s just much faster than the Linux version.
Hi Tim,
This is a comment, nor a line of code. After finishing the thread, the state will change and the following code is executed.
Marcelo
Dear Beatrix,
This is only a very small part of my software. Only an exemple of a thread and methods. The question is why processing in Windows is 10 times faster than in Linux.
Marcelo
That was my point. If you have a tight loop like that which is basically a while … do nothing … wend, it’s going to kill your performance. Try
da.run
while da.State = 0
app.SleepCurrentThread(250)
wend
It may considerably improve the performance of your app. Note: you can tweak the value of 250 to tune the performance.
Dear Jeff,
See bellow my reply for each one of your comments.
To me, a lot of time is likely being wasted with the redims
Redim is a method for releasing memory. My software can provide data for thousands of individuals and up to a million of SNPs.
It should be possible to keep a constant (large enough) array available, and just make a note about how much of it is used.
In simple terms, if you have a college with a potential 2000 stutends, your array could start at 2000. You record number_of_students - sometimes it is 1000, sometimes it is 1700, but the array never changes. That does mean there can be unused data ‘above the number’, but as long as you stop at the right spot, you will be fine. Not redimming means no constant reallocation of memory.
I am not sure how to replay this comment. The dimensions in my arrays depends on the user inputs for chromosomes, genes, DNA markers, number of parents, number os generations etc
Without comments, it is really hard to follow what this code does - it looks like it has grown organically, with IF statements added to cater for edge cases during testing.
The code is used for crossing a number of individuals between themselves. Then I get genes and markers in up to 10 chromosomes for each male and female pair.
The code shown is not optimised… adding the #pragma statements would help, but the truly slow part could be in any of the methods that this calls. eg gamfromGen
This takes gametes from a male or female.
Maybe the SaveData part can be optimised… each time that is called, you might open a file on disc and write to it.
Savedata is a method for save the output. Genotypes for DNA polymorphisms.
Changing that to ‘add a line to an in-memory string’, then writing the whole lot at the end might improve things, depending upon the size of the final data file.
Saving on disc in Linux is as fast as saving in Windows.
Marcelo
Of course. But the smaller the methods the better you can do profiling. Writing such large methods usually is a bad idea.